Health Data & Bioinformatics
Arbeitsgruppe
Inhalte
Die Arbeitsgruppe für Health Data and Bioinformatics am Institut für Medizintechnik und Informationsverarbeitung in Koblenz beschäftigt sich mit der Schnittstelle zwischen Medizintechnik, Künstlicher Intelligenz und datengetriebenen Gesundheitslösungen. Unser Ziel ist es, durch den Einsatz fortschrittlicher Datenanalyse, maschinellen Lernens und Bioinformatik die Patientenversorgung zu verbessern, klinische Entscheidungsprozesse zu optimieren und die personalisierte Medizin voranzutreiben.
Ein zentraler Schwerpunkt unserer Forschung liegt in der Entwicklung und Validierung von KI-Modellen für medizinische Anwendungen, wobei wir besonderen Wert auf deren Zuverlässigkeit, Transparenz und Integration in klinische Abläufe legen. Wir analysieren komplexe biomedizinische Daten, darunter elektronische Gesundheitsakten, medizinische Bildgebung und Genomdaten, um relevante Erkenntnisse für die Praxis zu gewinnen. In enger Zusammenarbeit mit Krankenhäusern, Forschungseinrichtungen und Industriepartnern setzen wir unsere Forschungsergebnisse in anwendbare Lösungen für das Gesundheitswesen um.
Durch interdisziplinäre Zusammenarbeit und methodische Innovation tragen wir zur Weiterentwicklung der Medizintechnik und der digitalen Gesundheit bei. Unser Ziel ist es, durch innovative Technologien die Gesundheitsversorgung zu verbessern und die digitale Transformation der Medizin voranzutreiben.
Projekte
JProf. Dr. Marie-T. Hopp
Characterization of the molecular basis of labile heme as a prothrombotic modulator under hemolytic conditions, Deutsche Forschungsgemeinschaft (DFG), 2022-2026.
Exploring the molecular basis of labile heme as a prothrombotic modulator in hemolytic disorders, Gesellschaft für Thrombose- und Hämostaseforschung (GTH), 2022-2025.
Dr. Sabine Bauer
Automatisierte Modellgenerierung der Halswirbelsäule mittels RGB-D Bilddaten für die Simulation und Analyse der Belastung von zervikalen muskoloskelettalen Strukturen (1.7.2021 bis zum 30.06.2024)
Prof. Dr. Peer Neubert
AI-DPA "Analyse und Interpretation von unstrukturierten Daten und Prozessen in zwei- und dreidimensionalen Anwendungsszenarien mit Machine Learning“ KI-Forschungskolleg in cooperation with University of Applied Science Mainz. (2023-2026)
MineSweeper: A research study on visual perception and optimization for localization and mapping in challenging and changing environments in the context of civil demining in cooperation with the BAAINBw. (2024-2027)
ViPRICE-2: A research project with the German Aerospace Center (DLR) and TU Chemnitz on Visual Place Recognition in Changing Environments. (2022-2025)
Prof. Dr. Maik Kschischo
DFG Research Unit 2800 (FOR2800): Chromosome Instability: „Cross-talk of DNA replication stress and mitotic dysfunction”, Sub-project SP-3: Modelling the link between replication stress, chromosomal instability and aneuploidy. Deutsche Forschungsgemeinschaft (DFG), 2019-2025.
Data2Health, Vertauenswürdige Datenanalysen im Gesundheitswesen, Forschungskolleg der Universität Koblenz und der Hochschule Koblenz, 2022-2025. Ministerium für Wissenschaft und Gesundheit Rheinladd-Pfalz und Debeka.
SEEDS-Structural Error Estimation in Dynamic Systems. Deutsche Forschungsgemeinschaft (DFG), 2017-2024.
TelemonKI, Kooperation mit der Qurasoft GmbH Koblenz, Entwicklung einer Software basierend auf kausalen Neuronal Ordinary Differential Equations (NODEs) für das Telemonitoring von Patienten mit chronischen Krankheiten, um kritische Ereignisse frühzeitig vorherzusagen und die Patientenversorgung zu verbessern. BMBF, 2024-2026
Publikationen
Böhly, N., Schmidt, A.-K., Zhang, X., Slusarenko, B. O., Hennecke, M., Kschischo, M., & Bastians, H. (2022). Increased replication origin firing links replication stress to whole chromosomal instability in human cancer. Cell Reports, 41(11). https://doi.org/10.1016/j.celrep.2022.111836
Chávez, J. P., Götz, T., Siegmund, S., & Wijaya, K. P. (2017). An SIR-Dengue transmission model with seasonal effects and impulsive control. Mathematical Biosciences, 289, 1–15. https://doi.org/10.1016/j.mbs.2017.04.005
Dönges, P., Götz, T., Kruchinia, N., Krüger, T., Niedzielewski, K., Priesemann, V., & Schäfer, M. (2024). SIR model for households. SIAM Journal of Applied Mathematics, 84(4). https://doi.org/10.1137/23M1556861
Götz, T., Krüger, T., Niedzielewski, K., Pestow, R., Schäfer, M., & Schneider, J. (2024). Chaos in opinion-driven disease dynamics. Entropy, 26(4). https://doi.org/10.3390/e26040298
Heidrich, P., Jayathunga, Y., Bock, W., & Götz, T. (2020). Prediction of dengue cases based on human mobility and seasonality—An example for the city of Jakarta. Mathematical Methods in the Applied Sciences, 26. https://doi.org/10.1002/mma.7648
Hopp, M.-T., Alhanafi, N., George, A. A. P., Hamedani, N. S., Biswas, A., Oldenburg, J., Pötzsch, B., & Imhof, D. (2021). Molecular insights and functional consequences of the interaction of heme with activated protein C. Antioxidants & Redox Signaling, 34(1), 32. https://doi.org/10.1089/ars.2019.799
Hopp, M.-T., Holze, J., Lauber, F., Holtkamp, L., Rathod, D. C., Miteva, M. A., Prestes, E. B., Manoury, B., Merle, N., Roumenina, L. T., Bozza, M. T., Weindl, G., & Imhof, D. (2024). Insights into the molecular basis and mechanism of heme-triggered TLR4 signaling: The role of heme-binding motifs in TLR4 and MD2. Immunology, 7(2), 181. https://doi.org/10.1111/imm.13708
Hopp, M.-T., Rathod, D. C., & Imhof, D. (2022). Host and viral proteins involved in SARS-CoV-2 infection differentially bind heme. Protein Science, 31(11), e3341. https://doi.org/10.1002/pro.4451
Hopp, M.-T., Ugurlar, D., Pezeshkpoor, B., Biswas, A., Ramoji, A., Neugebauer, U., Oldenburg, J., & Imhof, D. (2024). In-depth structure-function profiling of the complex formation between coagulation factor VIII and heme. Thrombosis Research, 237, 184. https://doi.org/10.1016/j.thromres.2024.04.006
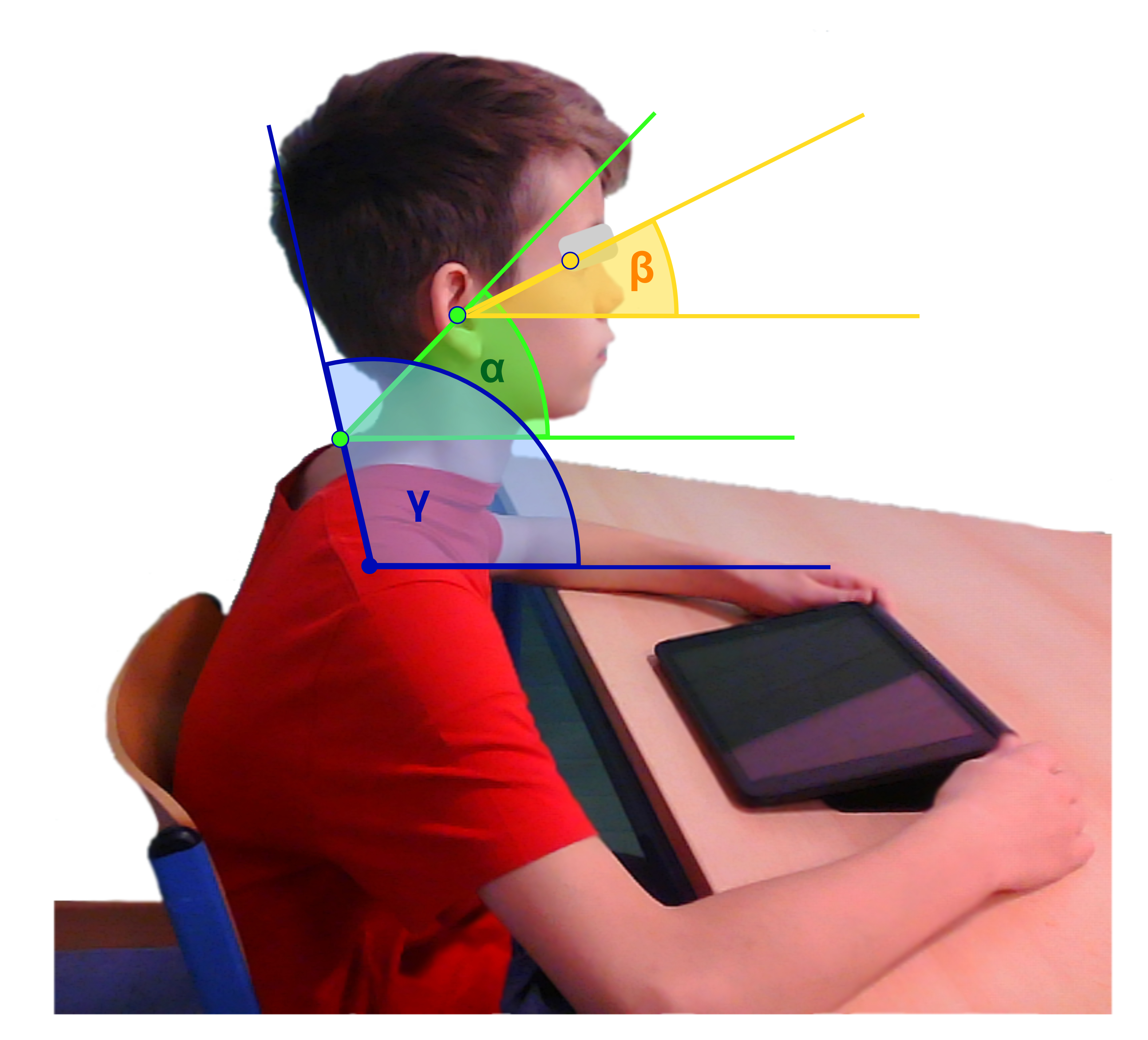
Kramer, I., & Bauer, S. (2023). Analysis of adolescents’ head to shoulder region during tablet use from sagittal and frontal RGB images. Applied Biosciences, 2(3), 421–436. https://doi.org/10.3390/applbiosci2030027
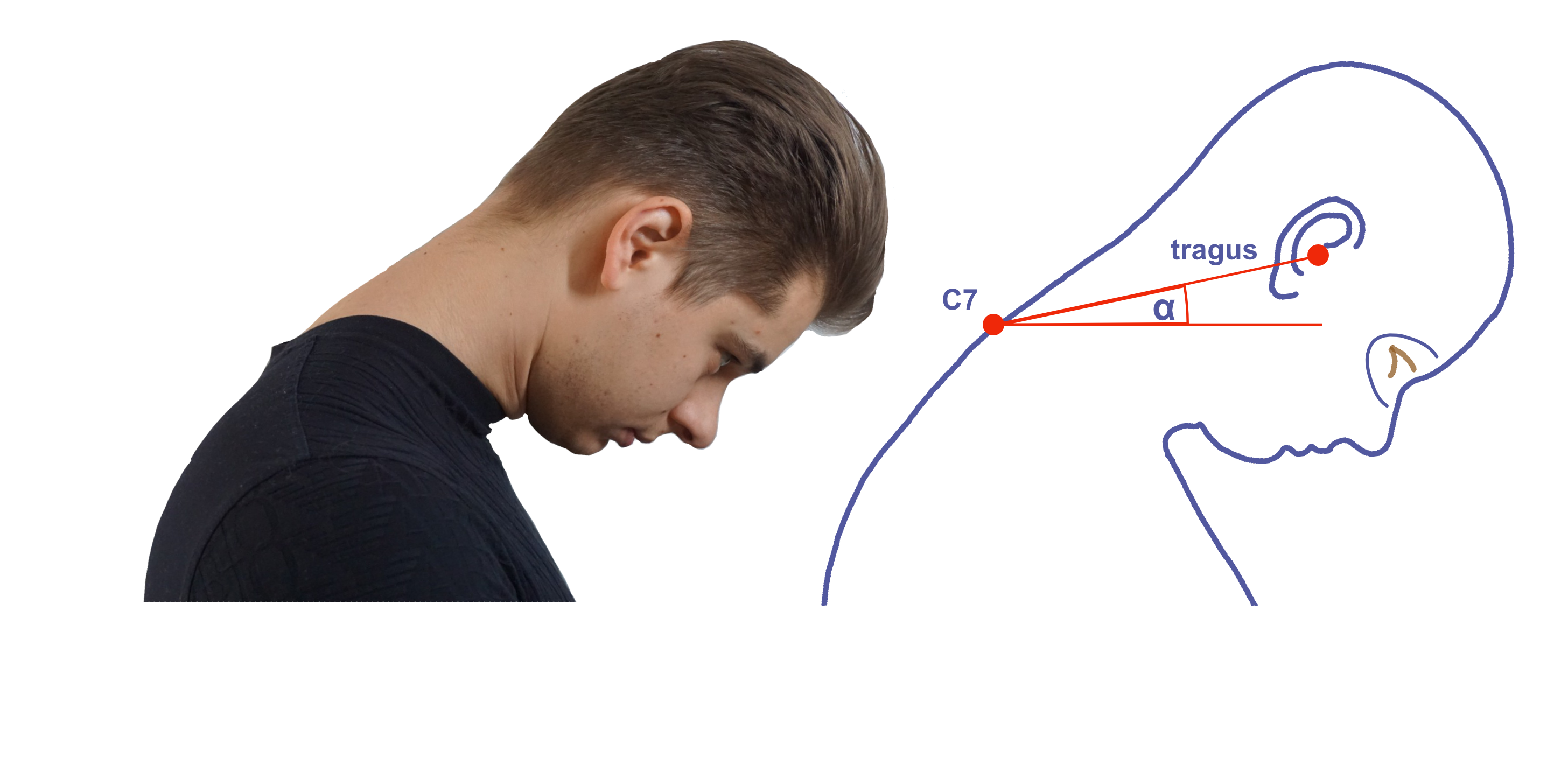
Kramer, I., Bauer, S., & Matejcek, A. (2022). Automated detection of ear tragus and C7 spinous process in a single RGB image: A novel effective approach. BioMedInformatics, 2(2), 318–331. https://doi.org/10.3390/biomedinformatics2020020
Kramer, I., Bauer, S., & Paulus, D. (2022). Holistic approach for markerless head-neck posture assessment. Journal of Head Neck & Spine Surgery, 4(5), 555646–555646. https://doi.org/10.19080/JHNSS.2022.04.555646
Mubeen, S., Domingo-Fernandez, D., Díaz del Ser, S., Solanki, D., Kodamulli, A. T., Hofmann-Apitius, M., Hopp, M.-T., & Imhof, D. (2022). Exploring the complex network of heme-triggered effects on the blood coagulation system. Journal of Clinical Medicine, 11(19), 5975. https://doi.org/10.3390/jcm11195975
Neubert, P., & Schubert, S. (2021). Hyperdimensional computing as a framework for systematic aggregation of image descriptors. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). https://doi.org/10.1109/CVPR46437.2021.01666
Neubert, P., Schubert, S., Schlegel, K., & Protzel, P. (2021). Vector semantic representations as descriptors for visual place recognition. In Proceedings of Robotics: Science and Systems (RSS). https://doi.org/10.15607/RSS.2021.XVII.083
Schäfer, M., Heidrich, P., & Götz, T. (2023). Modelling the spatial spread of COVID-19 in a German district using a diffusion model. Mathematical Biosciences and Engineering, 20(12), 21246–21266. https://doi.org/10.3934/mbe.2023940
Schlegel, K., Kleyko, D., Brinkmann, B. H., Nurse, E. S., Gayler, R. W., & Neubert, P. (2024). Lessons from a challenge on forecasting epileptic seizures from non-cerebral signals. Nature Machine Intelligence Challenge Accepted.
Schlegel, K., Neubert, P., & Protzel, P. (2022). A comparison of vector symbolic architectures. Artificial Intelligence Review. https://doi.org/10.1007/s10462-021-10110-3
Schubert, S., Neubert, P., Garg, S., Milford, M., & Fischer, T. (2023). Visual place recognition: A tutorial. IEEE Robotics and Automation Magazine (RAM). https://doi.org/10.1109/MRA.2023.3310859
Wendland, P., Schenkel-Häger, C., Wenningmann, I., & Kschischo, M. (2024). OptAB - an optimal antibiotic selection framework for Sepsis patients using artificial intelligence. Preprint, under review in npj Digital Medicine. https://doi.org/10.21203/rs.3.rs-4598166/v1
Wendland, P., Schmitt, V., Zimmermann, J., Häger, L., Göpel, S., Schenkel-Häger, C., & Kschischo, M. (2023). Machine learning models for predicting severe COVID-19 outcomes in hospitals. Informatics in Medicine Unlocked, 37, 101188. https://doi.org/10.1016/j.imu.2023.101188
Wijaya, K. P., Ganegoda, N., Jayathunga, Y., Götz, T., Schäfer, M., & Heidrich, P. (2021). An epidemic model integrating direct and fomite transmission as well as household structure applied to COVID-19. Journal of Mathematics in Industry, 1–26. https://doi.org/10.1186/s13362-020-00097-x
Wijaya, K. P., Götz, T., & Soewono, E. (2014). An optimal control model of mosquito reduction management in a dengue endemic region. International Journal of Biomathematics, 7(5), 1450056. https://doi.org/10.1142/S1793524514500569
Zhang, X., & Kschischo, M. (2021). MFmap: A semi-supervised generative model matching cell lines to tumours and cancer subtypes. PLOS ONE, 16(12), e0261183. https://doi.org/10.1371/journal.pone.0261183
Zhang, X., & Kschischo, M. (2022). Distinct and common features of numerical and structural chromosomal instability across different cancer types. Cancers, 14(6), 1424. https://doi.org/10.3390/cancers14061424